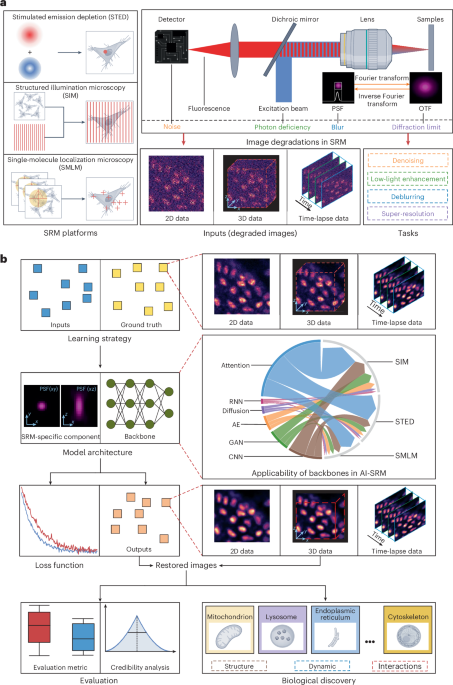
AI‑empowered super‑resolution microscopy (SRM) is rapidly becoming the cornerstone of modern cell biology, offering a level of detail that was once the exclusive domain of electron microscopy but with the added advantage of live‑cell compatibility. By coupling state‑of‑the‑art deep‑learning architectures with meticulously calibrated optical hardware, researchers can now capture images that reveal the intricate choreography of organelles, the fleeting interactions between protein complexes, and the subtle undulations of the cytoskeleton in real time. This synergy not only amplifies the visual fidelity of each frame but also streamlines the entire experimental pipeline: acquisition times are shortened, photobleaching is curtailed, and the massive data streams that once overwhelmed conventional storage solutions are processed on the fly by AI‑driven pipelines that prioritize relevant information and discard redundant noise. The result is a more sustainable, higher‑throughput approach to nanoscale imaging that empowers laboratories to pursue longer‑term studies of dynamic cellular processes without sacrificing sample integrity.
At the heart of this transformation are neural networks that have been trained to become expert “microscopists.” Convolutional models such as DnCNN and Noise2Noise have been fine‑tuned on thousands of paired low‑ and high‑signal microscopy frames, learning to differentiate genuine fluorescent signals from the stochastic fluctuations inherent to photon detection. These denoising networks can restore crisp detail from exposures that are orders of magnitude shorter than traditional methods would allow, opening the door to high‑speed tracking of vesicle trafficking or rapid calcium waves. Complementing denoising, generative adversarial networks like ESRGAN and its successors act as digital super‑samplers, inferring high‑frequency spatial information that lies beyond the diffraction limit by leveraging learned priors from extensive datasets of cellular structures. Because these generative models are trained on realistic ground‑truth images—often sourced from complementary techniques such as structured illumination or stimulated emission depletion microscopy—they can produce reconstructions that are not merely aesthetically pleasing but biologically faithful, preserving the quantitative relationships essential for accurate measurement.
Beyond image restoration, AI is reshaping the analytical workflow that follows acquisition. Vision transformers, with their ability to attend to global context across entire volumes, excel at segmenting densely packed organelles and tracing complex filament networks that would confound traditional threshold‑based algorithms. By integrating attention mechanisms, these models can maintain consistency across time series, allowing researchers to follow the lifecycle of individual mitochondria or the assembly of focal adhesions with minimal manual intervention. Unsupervised representation learning techniques, such as contrastive learning and clustering, are beginning to uncover previously invisible phenotypic variations in large unlabeled datasets, suggesting new avenues for hypothesis generation that do not rely on pre‑defined markers. This shift from a purely descriptive to a predictive framework is poised to accelerate discovery, enabling biologists to ask questions about cellular organization that were previously inaccessible due to the sheer complexity of the data.
The open‑source community has played a pivotal role in democratizing these advances. Tools like DeepSTORM provide end‑to‑end pipelines for single‑molecule localization that incorporate neural network‑based fitting, while the SR‑Microscopy Toolkit bundles a collection of pretrained models for denoising, super‑resolution, and segmentation, all packaged with easy‑to‑use interfaces for non‑expert users. Curated datasets such as BioSRM and CellularNanoNet offer high‑quality, multimodal images that span a wide range of cellular contexts, from bacterial substructures to mammalian nuclei, ensuring that models trained on these resources generalize robustly across experimental conditions. Community challenges like the OpenSRM competition not only benchmark the performance of emerging algorithms but also foster collaborative innovation, driving the field toward more transparent, reproducible, and scalable solutions.
Looking forward, the convergence of AI with SRM is set to blur the line between observation and experimentation. Real‑time adaptive control systems could enable microscopes to dynamically modulate laser intensity, exposure time, and focus based on instantaneous feedback from AI analyses, thereby optimizing the trade‑off between image quality and phototoxic stress for each individual cell. Multimodal integration platforms may soon fuse super‑resolution fluorescence maps with electron microscopy volumes or mass‑spectrometry‑derived chemical signatures, constructing comprehensive atlases that capture both structural and functional dimensions of cellular machinery. As hardware accelerators become more affordable and AI models more interpretable, the “AI everywhere” vision will transition from a distant promise to an everyday reality within the microscope’s optics, ushering in an era where nanoscale discovery is limited only by imagination rather than by the constraints of conventional imaging.